Visualizations with ggplot2
Bryan Mayer
2022-10-26
Getting Started
- Download the workshop materials to follow along:
- Unzip the folder.
- Load the project and open
ggplot2-worksheet-hvtn2022.Rmd- The presentation link is in the Rmd.
- Follow instructions in first chunk:
- Make sure you are in the correct directory.
ggplot2is the only required library for the exercises.- Load in data:
mock_bama_example
Code and Attribution
Many, many people have made ggplot2 tutorials!
Tidyverse help page: https://ggplot2.tidyverse.org/reference
Hadley Wickham’s text: R for Data Science
The presentation and code are publicly available on GitHub: https://github.com/bryanmayer/hvtn-ggplot2-workshop
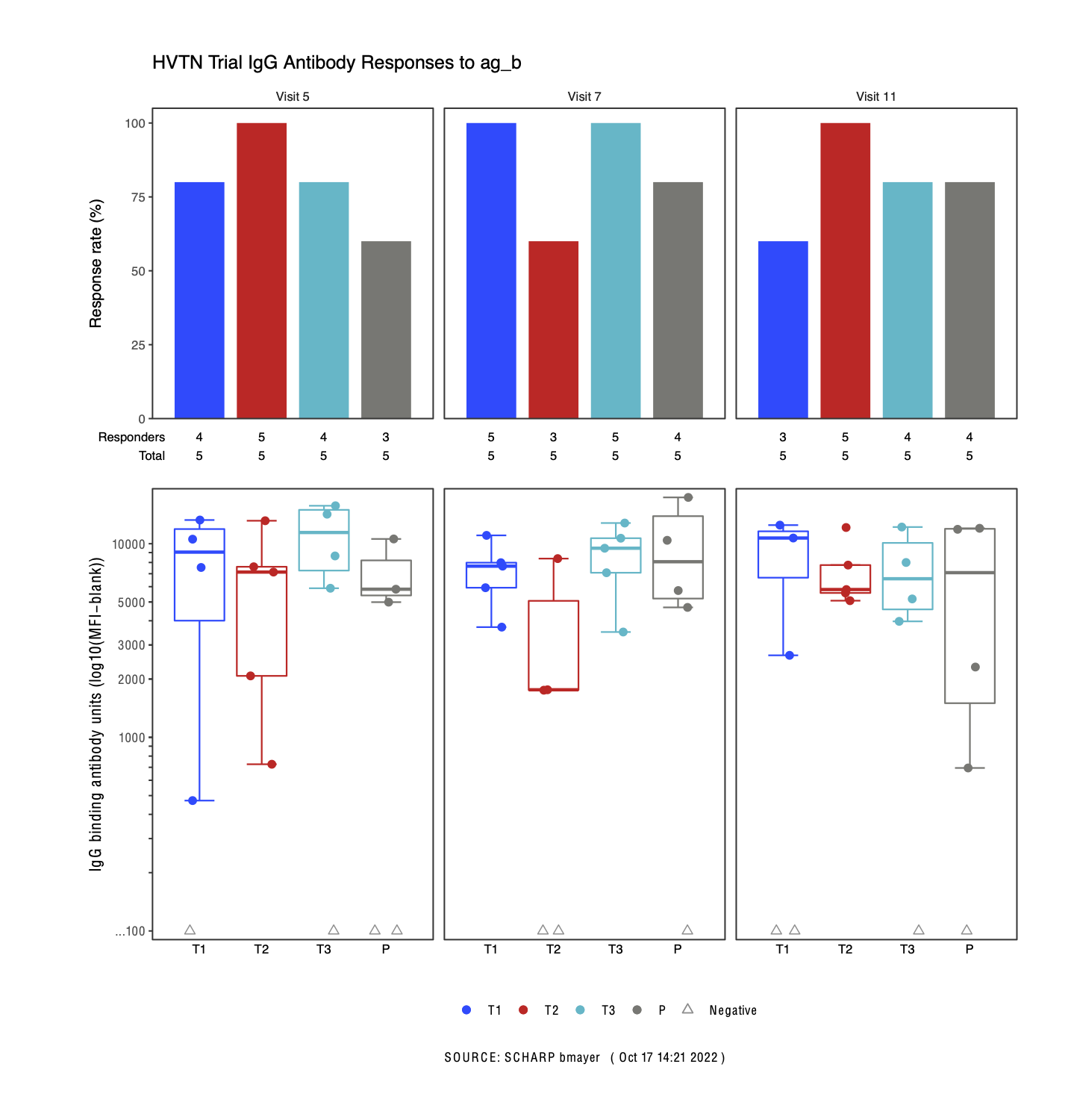
Goal today – Template HVTN figure
Roadmap
- ggplot2 basics
- Building ggplots with examples.
- Construct HVTN BAMA magnitude plot.
- If time, piece together full HVTN figure.
ggplot2 - the quintessential example
Basic template:
In action:
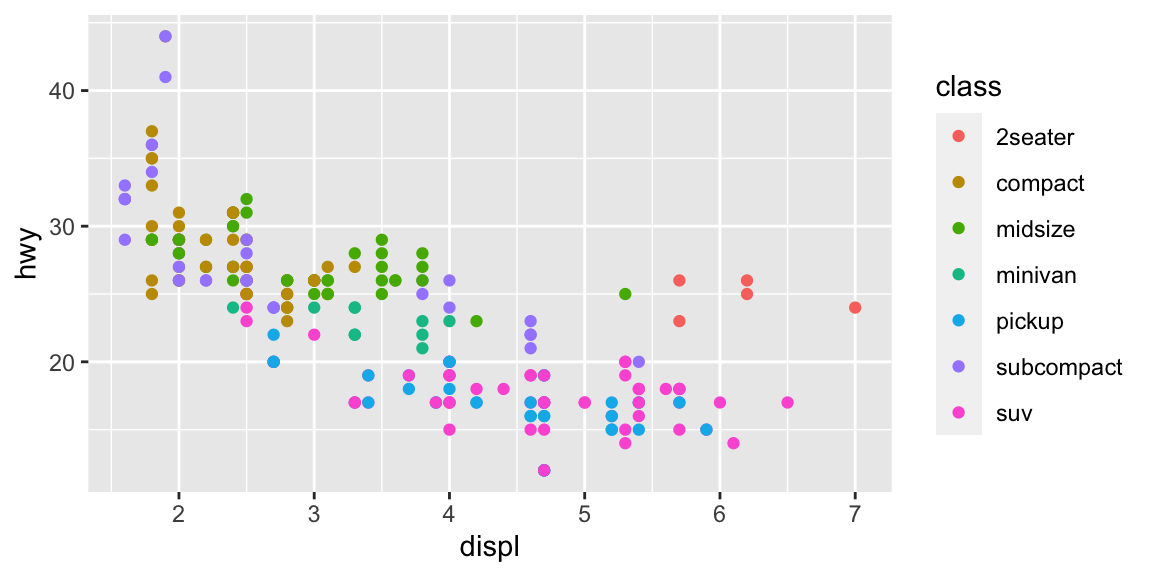
ggplot2 vs. base R plot
- For simple plots, base plot can be useful.
- Many R packages and objects use
plot()on the backend, so knowledge helpful.
- Many R packages and objects use
- The grammar of ggplot2 eases extension into complex plots.
- Ex. mapping of colors and legend generation is automatic within ggplot2.
par(mfrow = c(1,2))
plot(mpg$displ, mpg$hwy)
plot(x = mpg[mpg$class == "compact", ]$displ,
y =mpg[mpg$class == "compact", ]$hwy,
col = "orange", xlim = c(1.5, 7),
ylab = "hwy", xlab = "displ")
points(x = mpg[mpg$class == "2seater", ]$displ,
y = mpg[mpg$class == "2seater", ]$hwy,
col = "red")
legend(x = 3, y = 40, c("compact", "2seater"),
col = c("orange", "red"), pch = 1)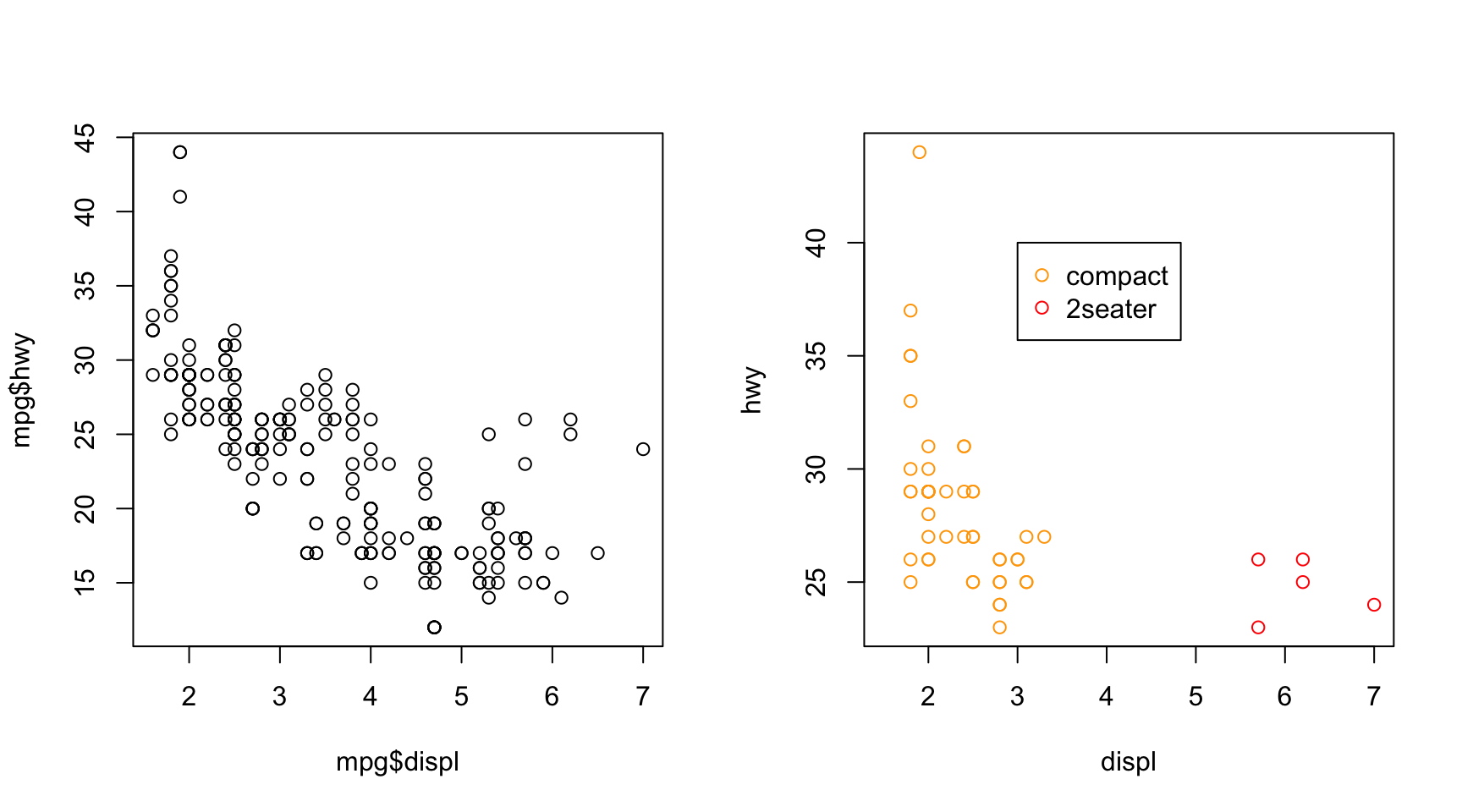
ggplot2 Basic Grammar
# explicitly stating 'mapping = aes()' is uncommon. "data = " necessary for geom assignments.
plA = ggplot(data = <DATA>, aes(<MAPPINGS>)) +
<GEOM_FUNCTION>()
plB = ggplot(aes(<MAPPINGS>)) +
<GEOM_FUNCTION>(data = <DATA>)
plC = ggplot(data = <DATA>) +
<GEOM_FUNCTION>(aes(<MAPPINGS>))
plD = ggplot() +
<GEOM_FUNCTION>(data = <DATA>, aes(<MAPPINGS>))ggplot()function initializes/creates a new plot.+is a ggplot2 operator (function) used to build a plot over function layers.<GEOM_FUNCTION>()(ex.geom_point()scatterplot) draws a geometry on the plot. I.e., ‘plot type’(s).dataargument for setting a data set andaes()function sets aesthetic mappings by variable.- Within
ggplot()sets global mappings across all plots. - Within
geom_**()will supersede global assignment but not apply to other geoms (local).
- Within
- ggplot2 plot objects can be saved as a variables.
- Layers can be added later to a ggplot2 object.
- Advanced: The internals of the ggplot2 object can be manipulated.
- Advanced: Multiple ggplot2 objects can be stored in a list (automation).
Common Geometries
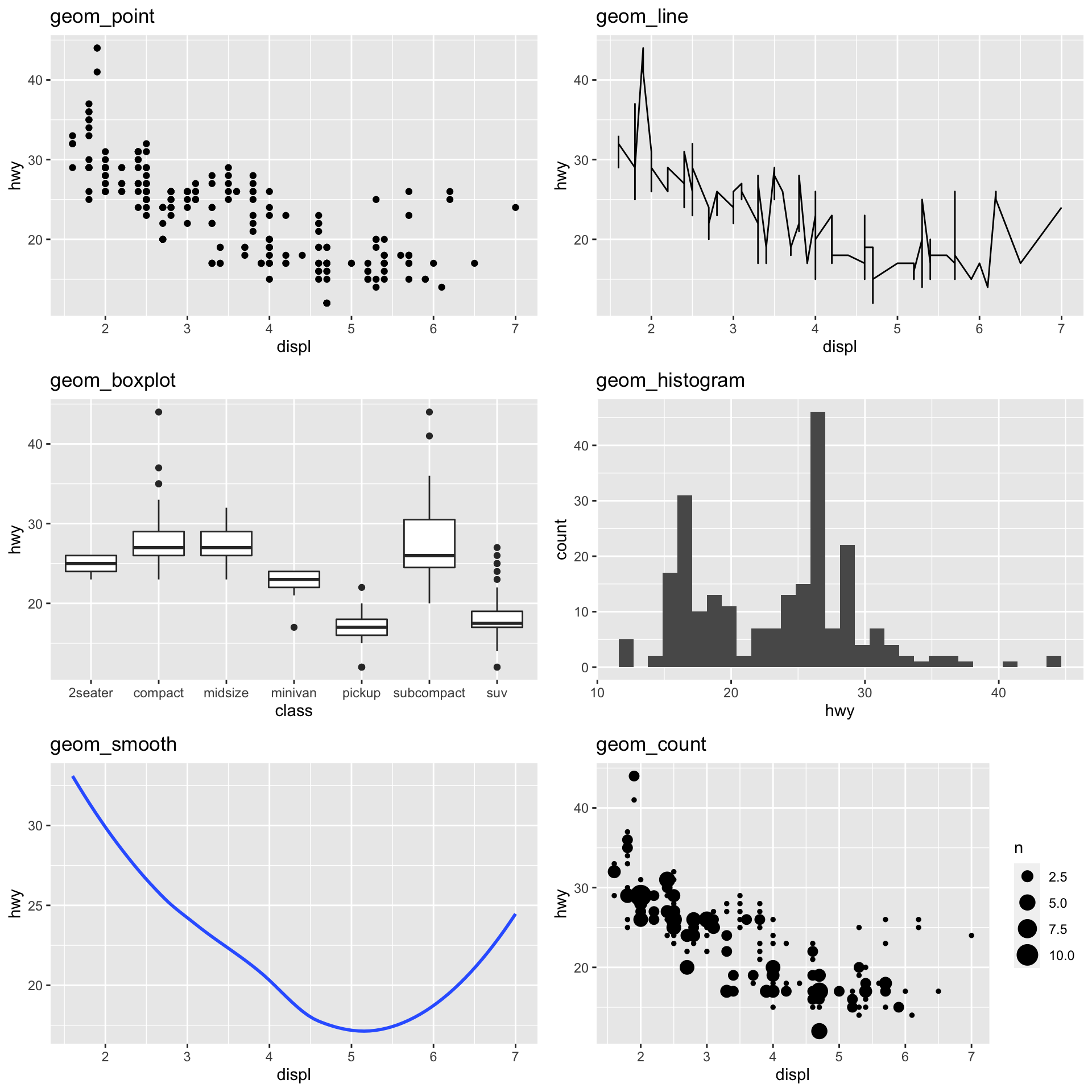
| Type | Function |
|---|---|
| Scatter/Point | geom_point() |
| Line | geom_line() |
| Box plot | geom_boxplot() |
| Bar plot | geom_bar(), geom_col() |
| Histogram | geom_histogram() |
| Density | geom_density() |
| Regression/Spline | geom_smooth() |
| Text | geom_text() |
| Vert./Horiz. Line | geom_{vh}line() |
| Jittered Point | geom_jitter() |
| Count | geom_count() |
https://eric.netlify.com/2017/08/10/most-popular-ggplot2-geoms/
Layering (combining) Geometries
Plots with multiple geometries can be quickly made by layering.
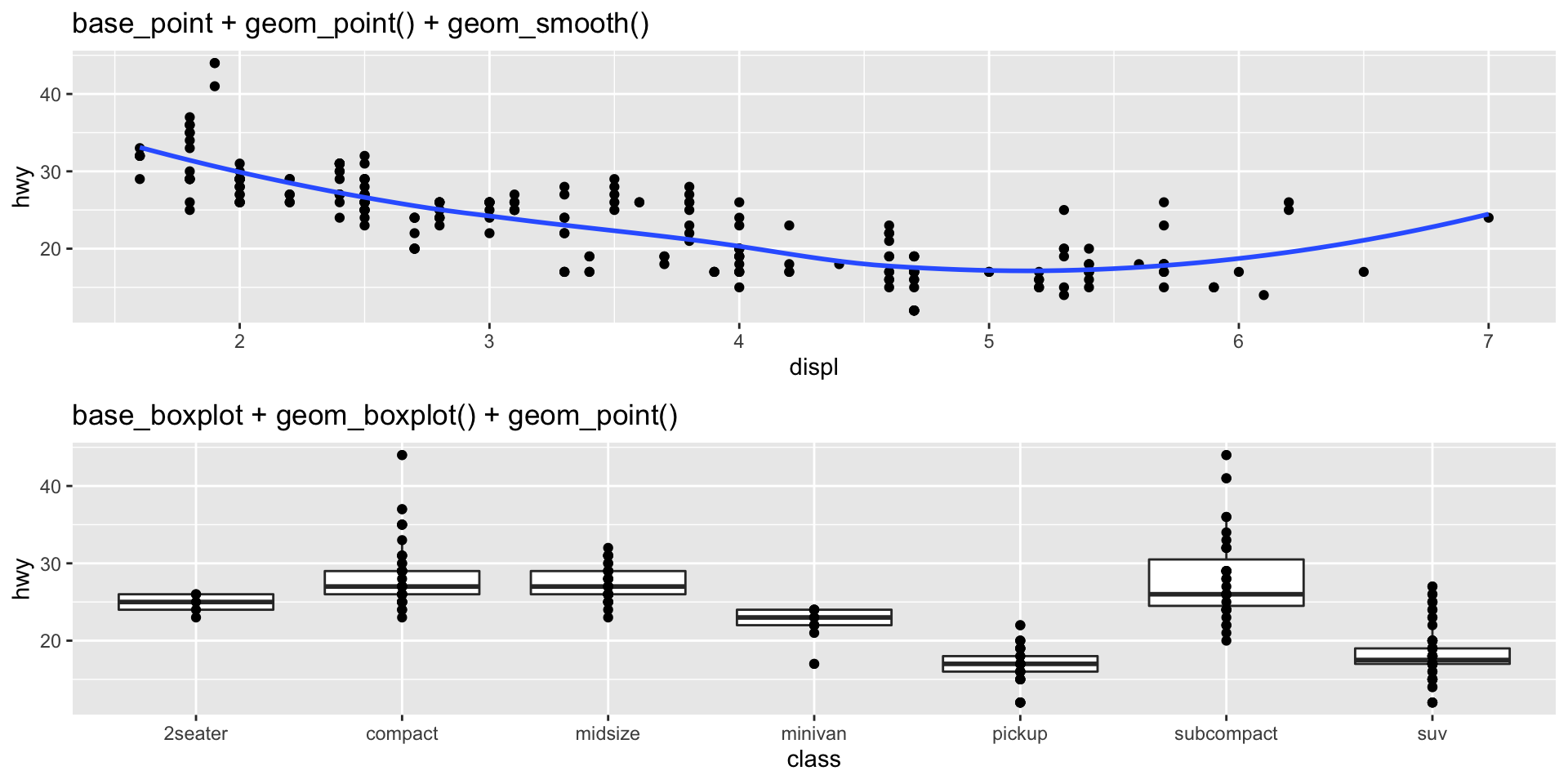
Aesthetic Mappings
Aesthetic mappings within
aes()link data to an aesthetic.Mappings depend on plot geometries.
- Check individual help files for information on them (ex.,
?geom_text). - Common source of error, but error message usually informative.
> base_boxplot + geom_histogram()Error in
f(): ! stat_bin() can only have an x or y aesthetic. Runrlang::last_error()to see where the error occurred.- Check individual help files for information on them (ex.,
Common Aesthetics
| Aesthetic | aes() Argument |
|---|---|
| x-axis | x |
| y-axis | y |
| Color outline | color |
| Color fill | fill |
| Point shape/type | shape |
| Point size | size |
| Opacity | alpha |
| Line type | linetype |
| Text | label |
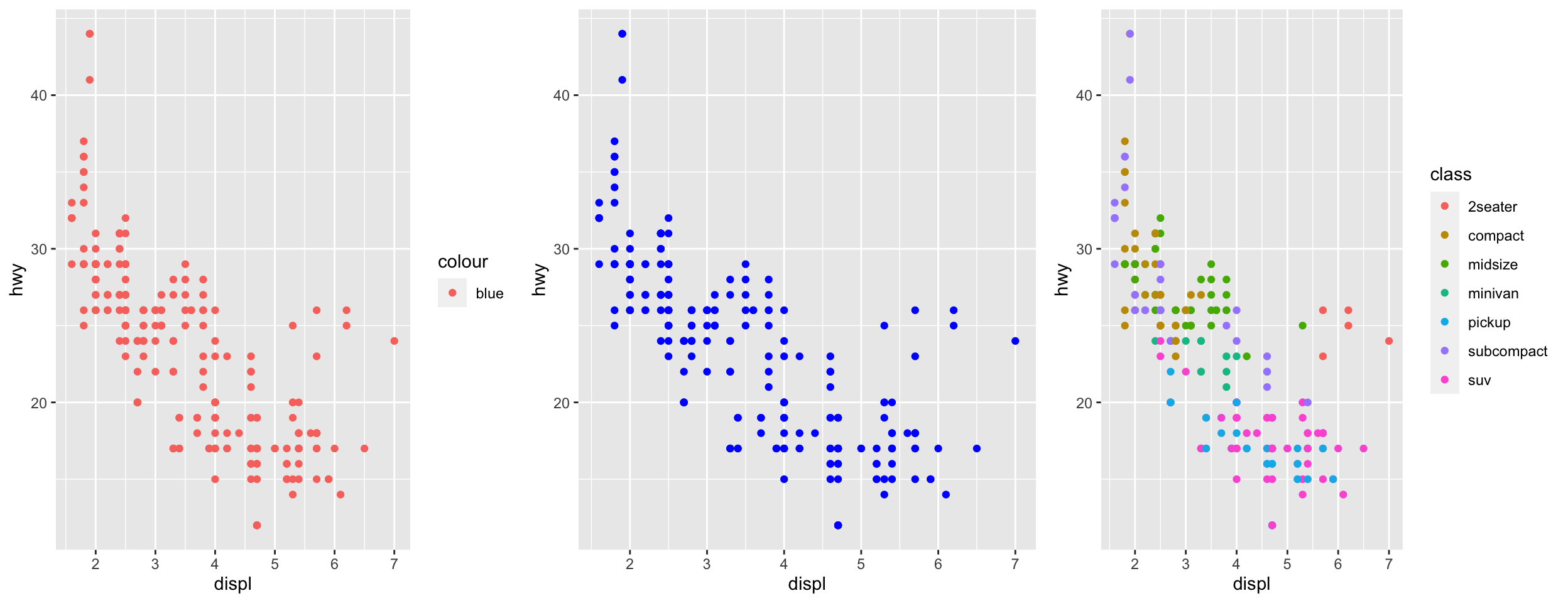
Aesthetic Settings vs. Mappings
- Setting: an aesthetic independent of data, assigned outside of the
aes()argument.- This must be assigned within the appropriate geometry.
- Assigning a setting within
aes()is a data mapping, ggplot2 will assume this a variable with a single value.
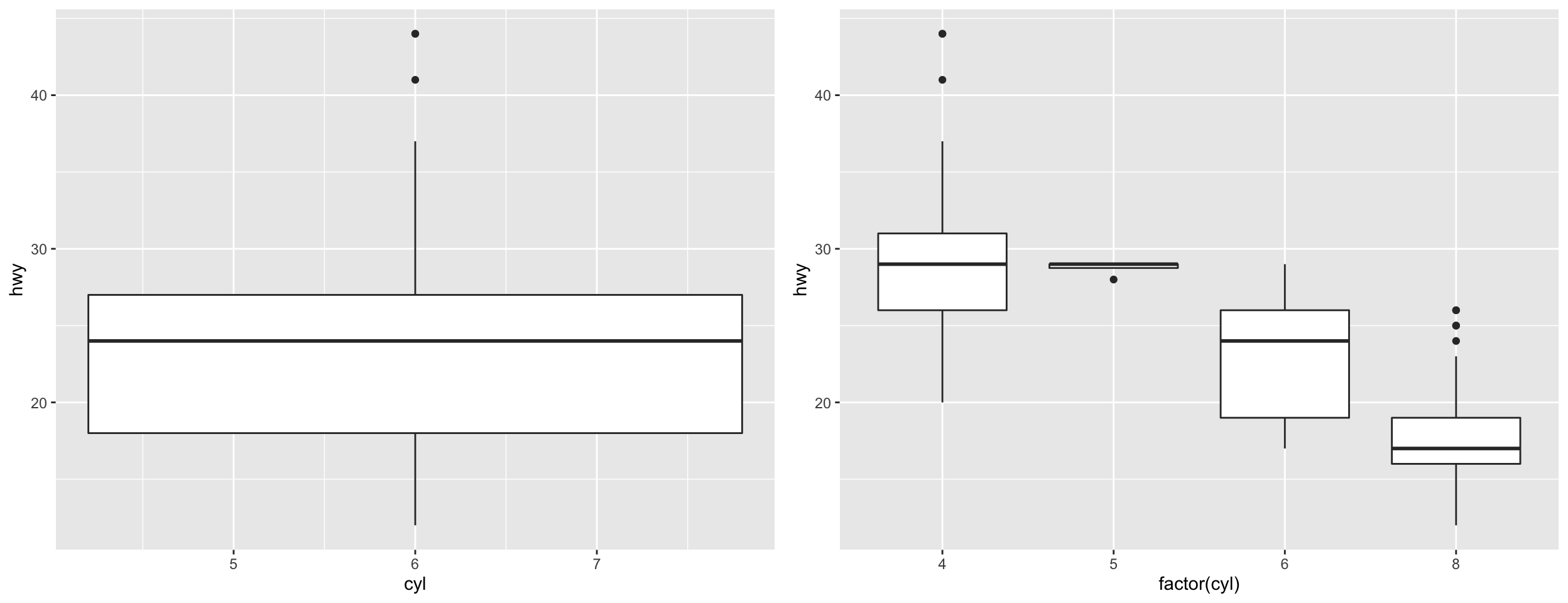
Continuous vs. Discrete Data
- Aesthetic mappings depend on type of data.
- ggplot2 will assume numeric classes are continuous.
- If the aesthetics requires a certain data, an informative error will display.
- Depending on geometry, aesthetics may only take one type of data (ex. linetype must be categorical class).
- If the aesthetic is flexible, the plot may appear odd (potentially with warning).
- If the aesthetics requires a certain data, an informative error will display.
- You can wrap variables with functions within
aes()calls.- Ex.
aes(y = log10(magnitude)),aes(y = pmax(1, magnitude)). - Trick: Wrap numeric variables in
factor()if need discrete mapping.
- Ex.
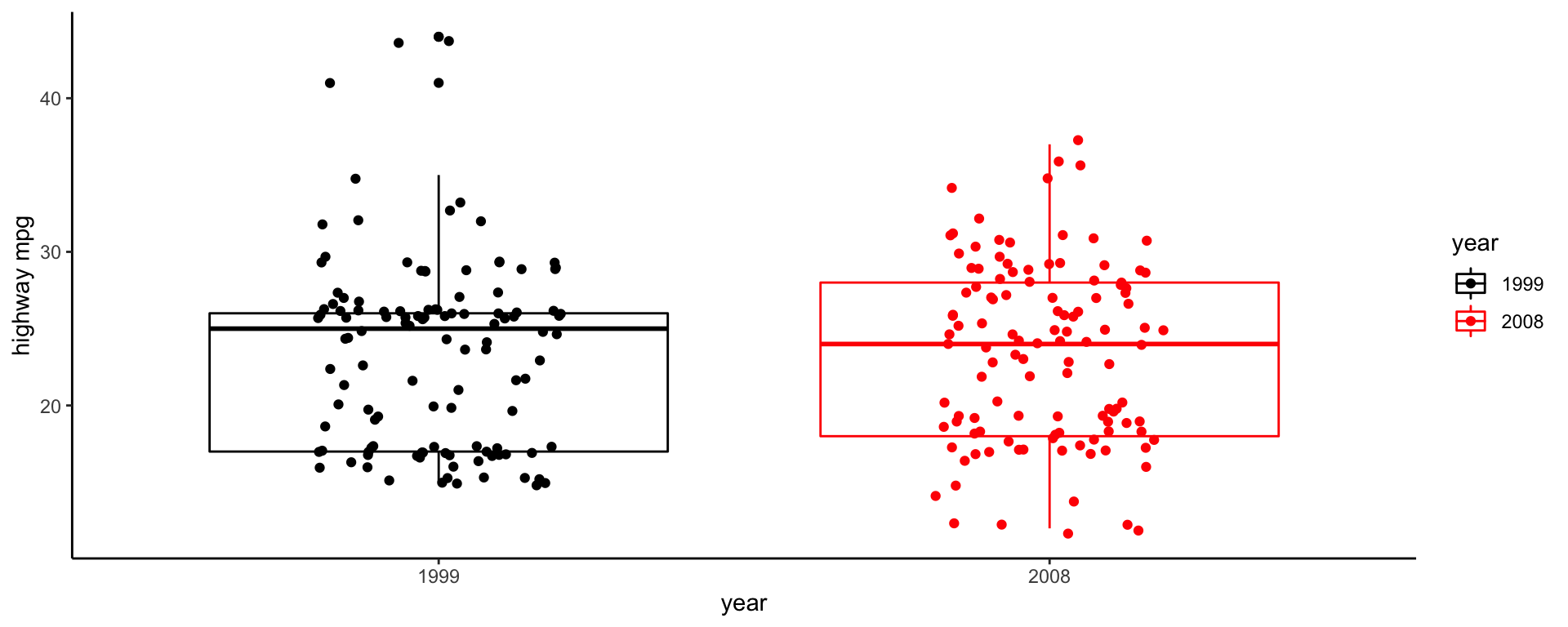
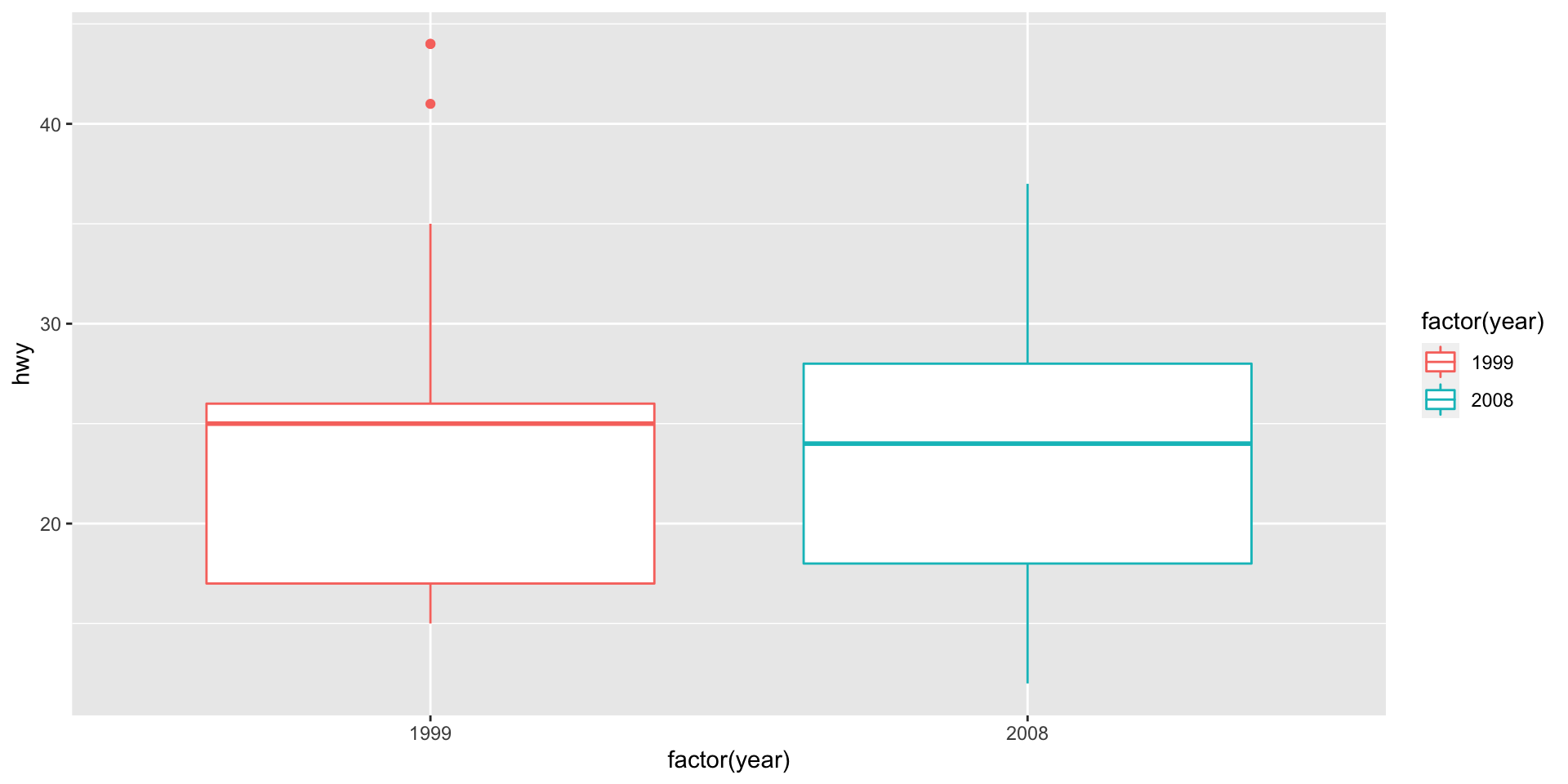
Boxplot example (mpg) - setup
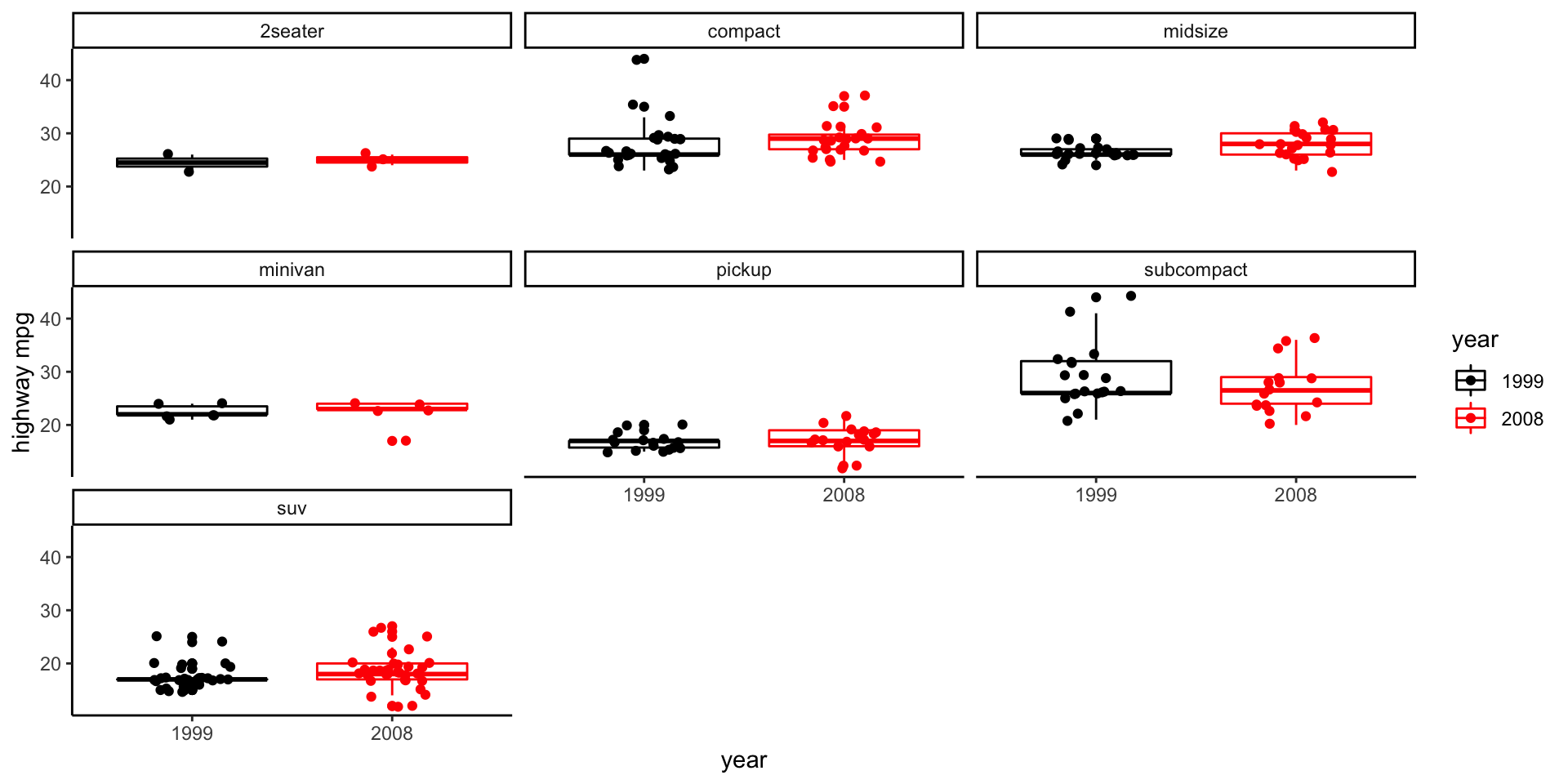
Question: Did highway mpg (hwy) improve between 1999 and 2008 (year)?
Data fields:
hwyyear(which is x?, which is y?)Geometry?
geom_boxplotMappings: let’s map year to color.
Take 5 minutes to make this plot. Work with your neighbor.
05:00
Box plot example (mpg) - v1
Why Factor?
Scale functions
- Scale functions: ‘transformation’ of the aesthetic mapping.
- Specific mapping for each level of the data (ex. level 1 <=> “green”).
- Scale function inputs:
- First argument (
name) is always the element (legend or axis) title. Default to variable name. - General arguments:
limits,breaks,labels- Customize legend and axes ticks and labels.
- A way to apply transformations to the data as they are plotted.
- First argument (
- Generating of high quality plots often requires manipulation of the scale functions.
- Types:
scale_*_discrete(),scale_*_manual(),scale_*_continuous()
Discrete and Manual Scale Functions
- Discrete scales (
scale_*_discrete()): tinkering with palettes, labels, and breaks.- Adjusting discrete x- or y-axis (
scale_x_discrete,scale_y_discrete) is not always intuitive (e.g., use limits instead of breaks).
- Adjusting discrete x- or y-axis (
- Manual scales (
scale_*_manual): explicit mapping of data level to a setting.- Make sure breaks and labels match as expected.
scale_color_manual(breaks = c("A", "B"), labels = c("A", "B"), values = c("red", "blue"))
Manual labeling
- Use named vectors for consistent repeated scale mappings.
- Ex. Same vaccine group color and label assignments across many plots.
- Note: Use ` ` for non-standard R column names.
- Not shown here: reference data frame (or list) linking breaks, labels, colors
breaks = dataframe$breaks, labels = dataframe$labels, values = dataframe$colors
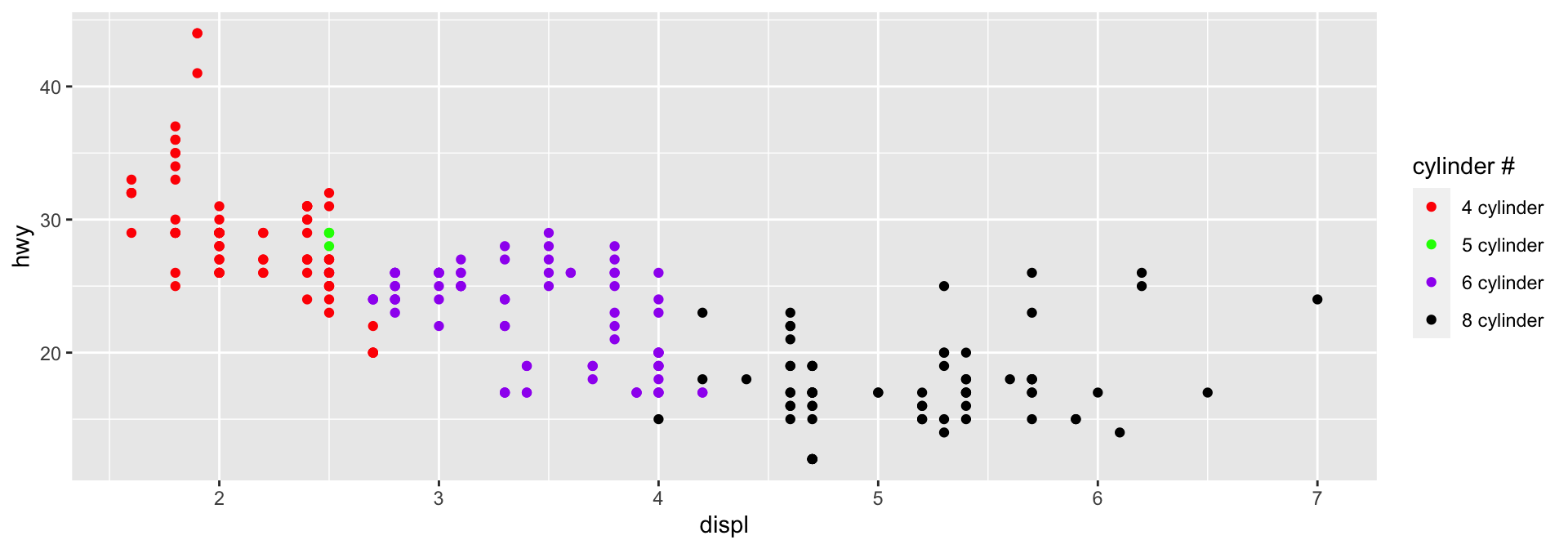
cyl_colors = c(`4` = "red", `5` = "green",
`6` = "purple", `8` = "black")
cyl_labels = paste(c(4, 5, 6, 8), "cylinder")
names(cyl_labels) = c(4, 5, 6, 8)
ggplot(data = mpg,
aes(x = displ, y = hwy,
colour = factor(cyl))) +
geom_point() +
scale_color_manual("cylinder #",
values = cyl_colors,
labels = cyl_labels)
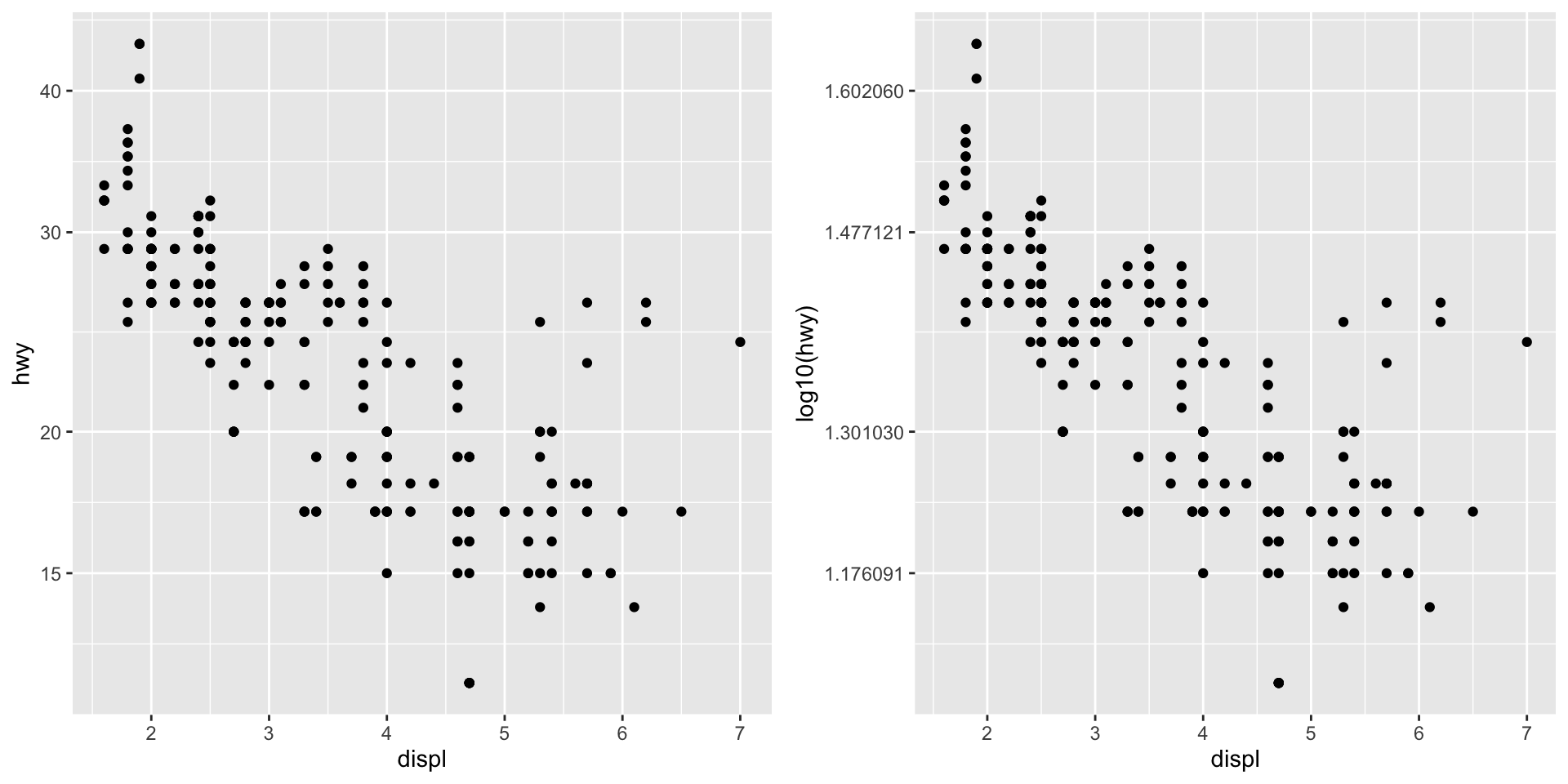
Continuous Scales
- Continuous scale manipulation tends to apply to continuous data mapped to axes.
- Common exceptions: heat maps, point size mapping.
- The default x- and y-axis scales are
scale_x_continuousandscale_y_continuous.- Most common alternative, log-transform:
scale_x_log10andscale_y_log10. - Plotting on the log-scale vs. log-transformed data (units are different).
- Other transformations are possible.
- Most common alternative, log-transform:
- More info: https://ggplot2.tidyverse.org/reference/index.html#section-scales
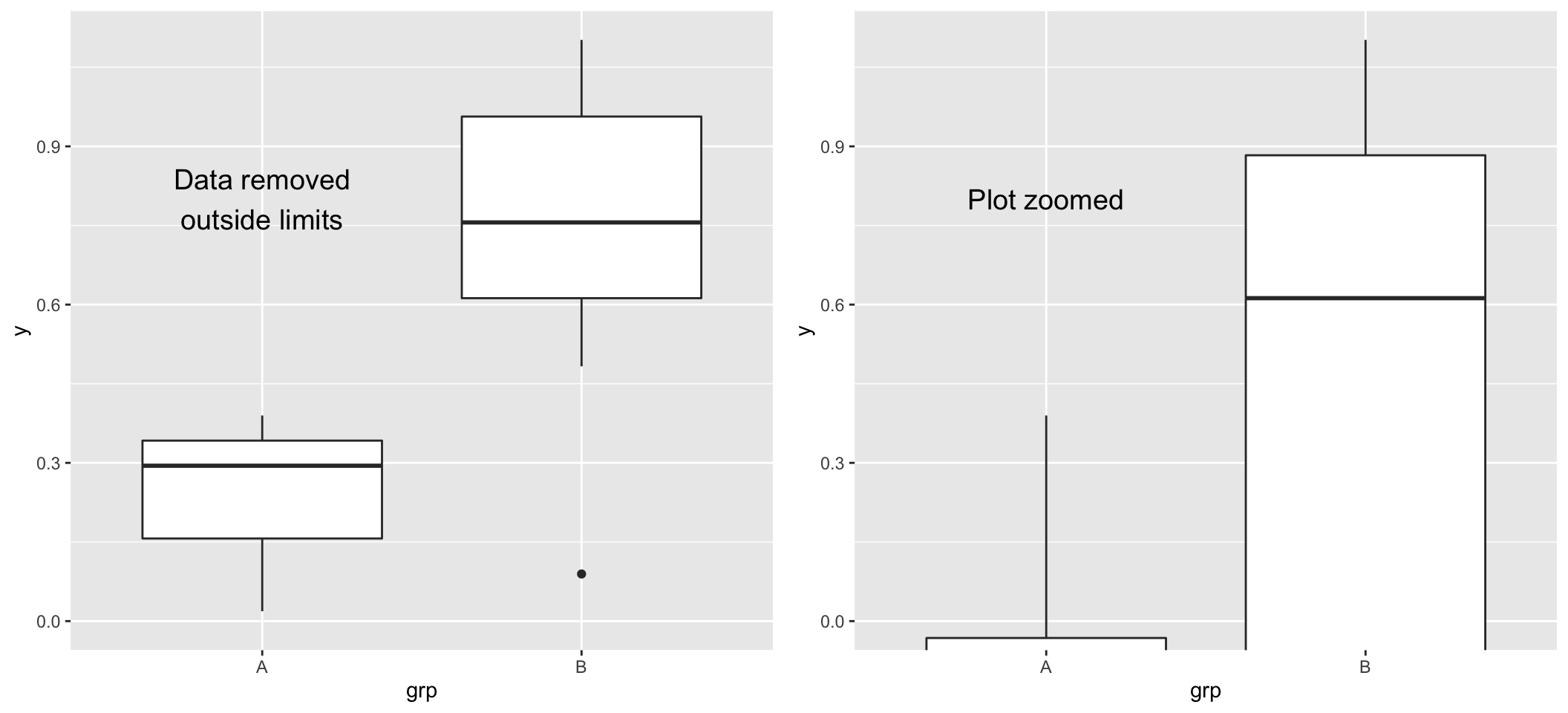
Coordinate functions
- Coordinate functions adjust the coordinate system of the plot.
- Not direct transformation to the data: different than scale mappings. Important when summary measures (e.g., median) are computed by ggplot2.
- Always read the warnings.
- Common coordinate functions:
coord_cartesian(): default. Limit arguments can be used to zoom plot (Example).coord_flip(): flips x and y on the plots.coord_polar(): use polar coordinate system.coord_fixed(): fixes aspect ratio, make ‘square’ plots.
Making a nice looking plot.
- Adjusted title and labels for axes and legends.
scale_y_continuous("y title", breaks = 5*3:9).- Set breaks and labels within the appropriate scale function.
- Another way to change legend and axes titles:
labs(x = "x title", y = "y title" color = "legend title")- Alternative axes title:
xlab(),ylab()
theme():- There are lots of plot settings to adjust (see
?theme). - General themes can be made and applied across multiple plots.
- ggplot2 also supplies themes:
theme_bw()theme_classic()
- ggplot2 also supplies themes:
- There are lots of plot settings to adjust (see
Box plot example (mpg) - Improve
Take 5 minutes. Work with your neighbor.
- Make the labels nicer.
- label x-axis as ‘year’, y-axis as ‘highway mpg’
- Map year to color.
- make 1998 = red, 2008 = black
- then 1998 = black, 2008 = red.
- Can you generalize this task?
- Show the raw data as points. (Bonus: jitter them.)
- Apply
+ theme_classic()
05:00
Box plot example (mpg) - finalize
Faceting
facet_gridorfacet_wrapcreate multiple panels across a variable level.facet_grid: structured, explicit mapping of rows and col (rectangular matrix).- 2-var ex.
+ facet_grid(rows = vars(var1), cols = vars(var2))or+ facet_grid(var1 ~ var2) - ex.
+ facet_grid(rows = vars(var1))or+ facet_grid(var1 ~ .) - ex.
+ facet_grid(cols = vars(var1))or+ facet_grid(. ~ var1)
- 2-var ex.
facet_wrap: sequence of panels across variable or layered variables.- ex.
+ facet_wrap(vars(var1))or+ facet_wrap(~ var1)
- ex.
- Last mpg exercise: use facteting to investigate whether 1998 vs. 2008 highway mpg varies by car
class.
Box plot example (mpg) - faceting
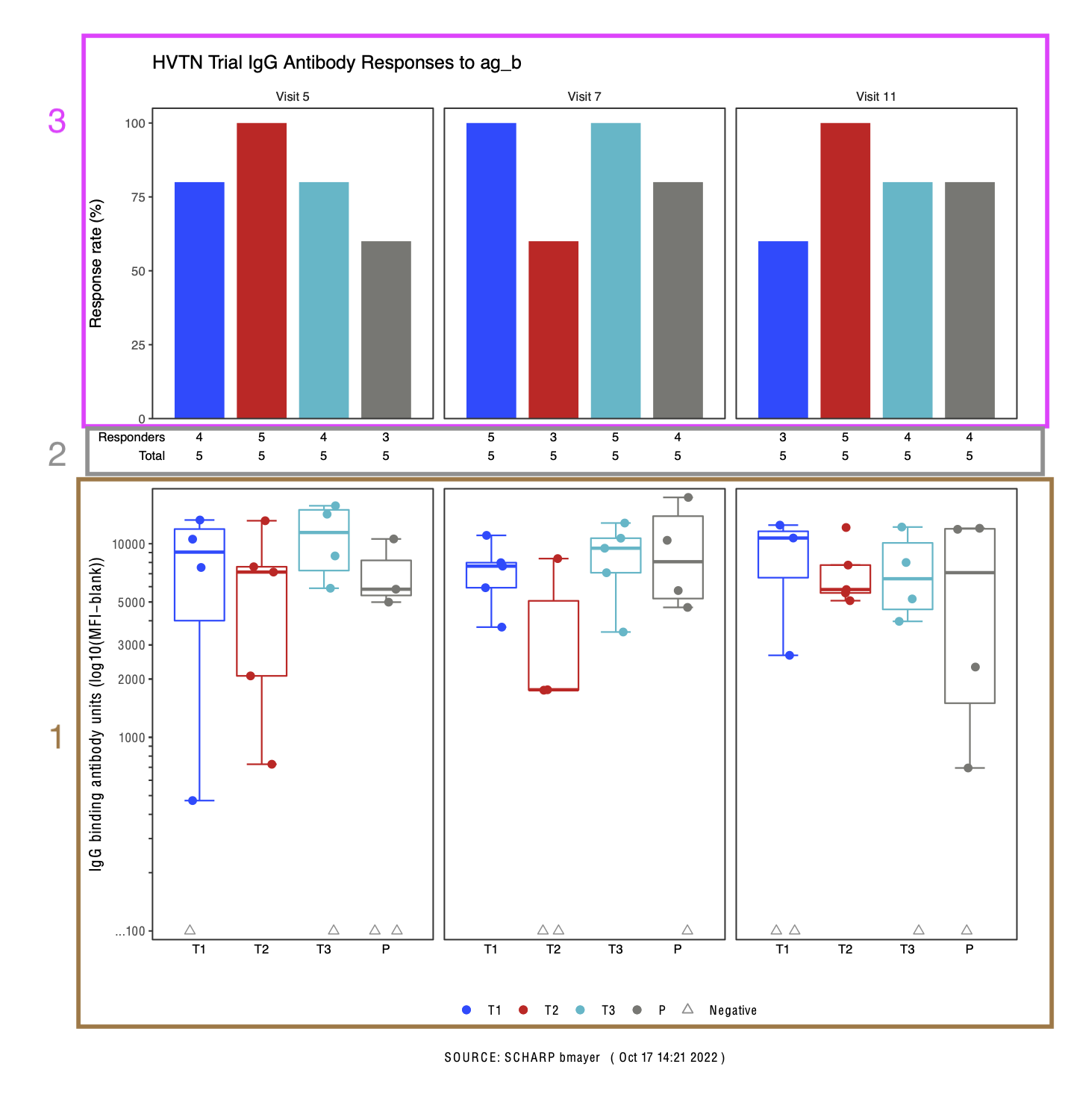
HVTN Example
Figure can be built in 3 parts. Focus just on magnitude plot.
HVTN - Data and Setup
Rows: 60
Columns: 12
$ labid <chr> "Frankenstein", "Frankenstein", "Frankenstein", "Franken…
$ pub_id <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 1…
$ rx_code <chr> "P", "T1", "T2", "T3", "P", "T1", "T2", "T3", "P", "T1",…
$ visitno <dbl> 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5,…
$ isotype <chr> "IgG", "IgG", "IgG", "IgG", "IgG", "IgG", "IgG", "IgG", …
$ antigen <chr> "ag_b", "ag_b", "ag_b", "ag_b", "ag_b", "ag_b", "ag_b", …
$ gene <chr> "gp120", "gp120", "gp120", "gp120", "gp120", "gp120", "g…
$ clade <chr> "B", "B", "B", "B", "B", "B", "B", "B", "B", "B", "B", "…
$ fi_bkgd <dbl> 11985.2477, 6188.5244, 6611.3519, 12819.1074, 5344.8246,…
$ fi_bkgd_blank <dbl> 6164.825, 6194.799, 4535.088, 4173.354, 5556.441, 6911.0…
$ delta <dbl> 5820.4230, 1.0000, 2076.2640, 8645.7529, 1.0000, 471.266…
$ response <dbl> 1, 0, 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1,…We have fixed colors and shapes, let’s assign those now:
Magnitude plot - Mapping (discussion)
Rows: 60
Columns: 12
$ labid <chr> "Frankenstein", "Frankenstein", "Frankenstein", "Franken…
$ pub_id <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 1…
$ rx_code <chr> "P", "T1", "T2", "T3", "P", "T1", "T2", "T3", "P", "T1",…
$ visitno <dbl> 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5,…
$ isotype <chr> "IgG", "IgG", "IgG", "IgG", "IgG", "IgG", "IgG", "IgG", …
$ antigen <chr> "ag_b", "ag_b", "ag_b", "ag_b", "ag_b", "ag_b", "ag_b", …
$ gene <chr> "gp120", "gp120", "gp120", "gp120", "gp120", "gp120", "g…
$ clade <chr> "B", "B", "B", "B", "B", "B", "B", "B", "B", "B", "B", "…
$ fi_bkgd <dbl> 11985.2477, 6188.5244, 6611.3519, 12819.1074, 5344.8246,…
$ fi_bkgd_blank <dbl> 6164.825, 6194.799, 4535.088, 4173.354, 5556.441, 6911.0…
$ delta <dbl> 5820.4230, 1.0000, 2076.2640, 8645.7529, 1.0000, 471.266…
$ response <dbl> 1, 0, 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1,…- Geometry?
- Aesthetic Data Mappings
- Which data fields are used in the plot?
- How are they mapped?
- Any transformations?
- Facets?
- Challenges?
Magnitude plot - Exercise
- Geometry:
geom_boxplot,geom_point(jitter) - Data fields: magnitude (
delta), groups (rx_code), response (response) - aes mappings: group to color,
responseto shape - Transformations: magnitude is truncated and plotted on the log10-scale.
- Facets: visits (
visitno) - Challenges:
- Separate color for ‘Negative’ within each group (and legend).
- Exclude non-responders from box plots.
- Exercise: make magnitude box plot from worksheet skeleton.
- “Exercise 3 - HVTN Magnitude plot” ~ line 221
- Hints provided on the worksheet in the plot skeleton.
- Ignore color issue for Negative.
Magnitude plot - Skeleton (Line 221)
ggplot(mock_bama_example, aes(...)) +
geom_boxplot(..., show.legend = F, outlier.colour = NA) +
geom_point(..., position = position_jitter(width = 0.2)) +
scale_shape_manual(values = c(2, 16)) +
scale_color_manual(values = hvtn_pl_colors) +
scale_x_discrete(limits = ...) +
scale_y_...() +
facet_wrap(...) +
labs(...) +
theme_classic() - Geometry:
geom_boxplot,geom_point(jitter) - Data fields: magnitude (
delta), groups (rx_code), response (response) - aes mappings: group to color,
responseto shape - Transformations: magnitude is truncated and plotted on the log10-scale.
- Facets: visits (
visitno) - Ignore negative responder color mapping.
05:00
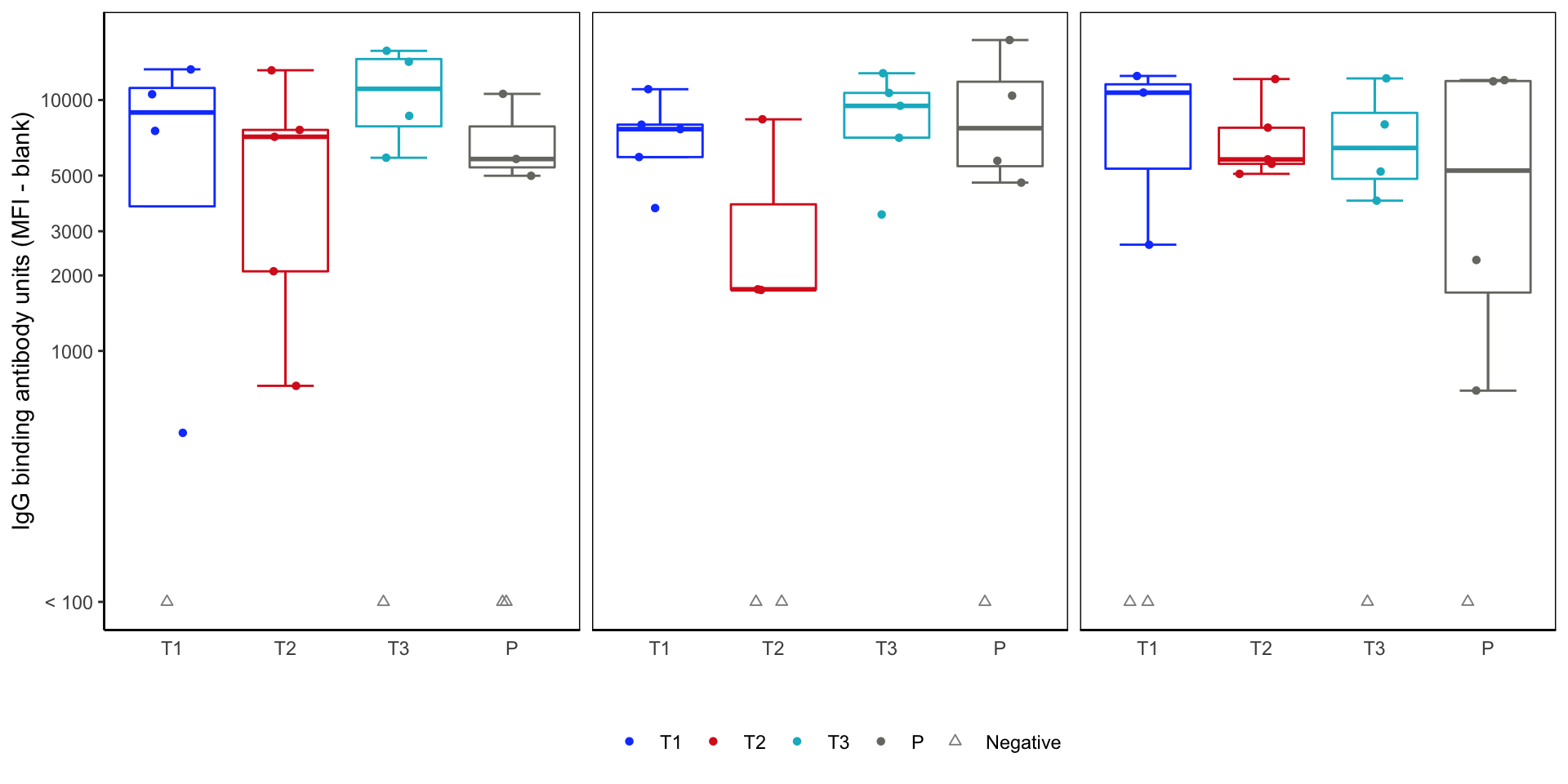
Response magnitude plot - v1
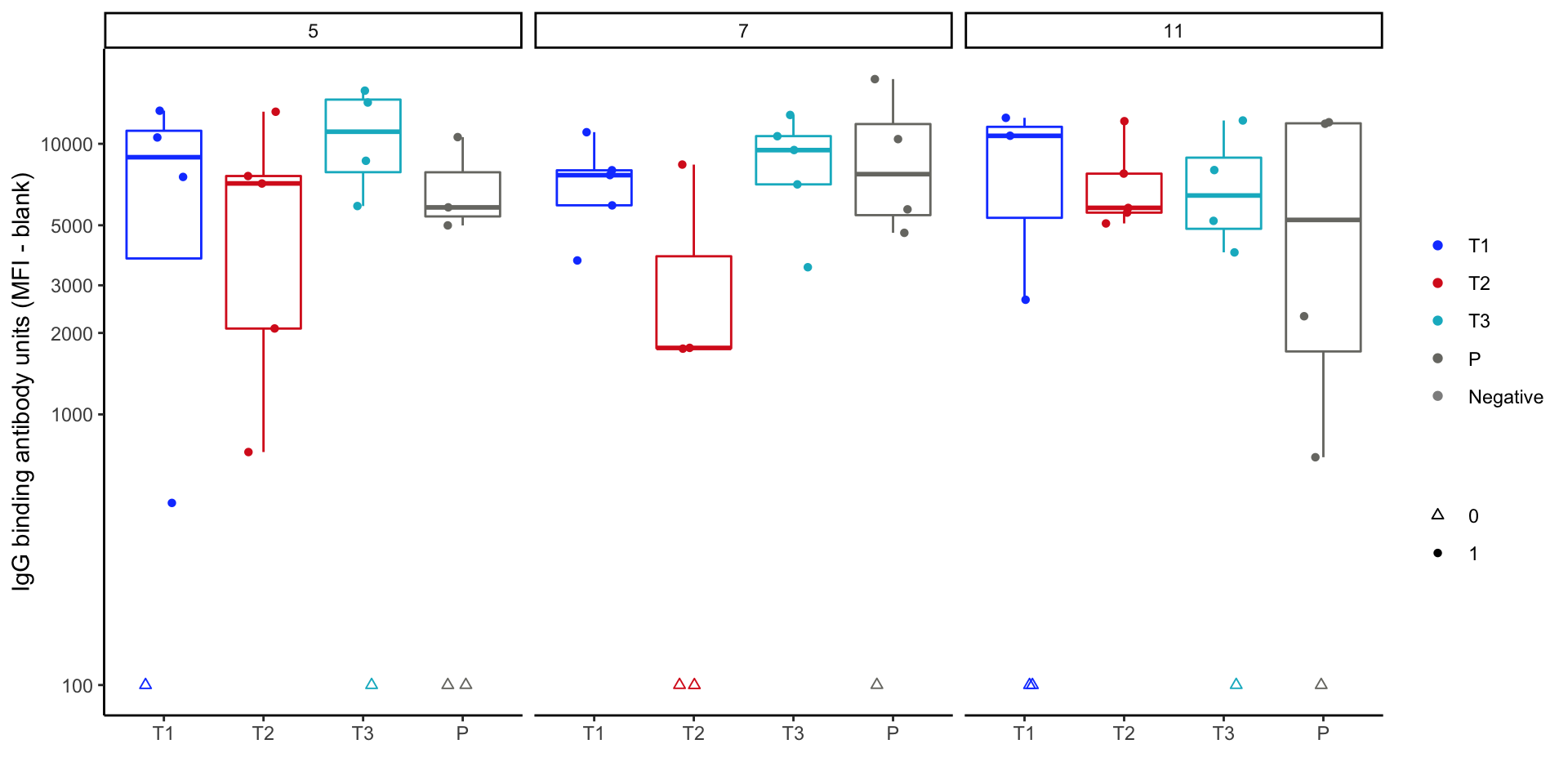
Response magnitude box plots - tinkering
- Rethink the mapping:
rx_codeto x-axis, but need new map to color.
- Update the plot using this variable for color mapping.
03:00
Magnitude plot - v2 Code
set.seed(14) # this keeps the jitter consistent each time
mag_bp_v2 = ggplot(data = mock_bama_example,
aes(x = rx_code, y = pmax(100, delta), color = grp_response)) +
geom_boxplot(data = subset(mock_bama_example, response == 1),
show.legend = F, outlier.colour = NA) +
geom_point(aes(shape = grp_response, group = rx_code),
position = position_jitter(width = 0.2)) +
scale_colour_manual("", values = hvtn_pl_colors) +
scale_shape_manual("", values = hvtn_pl_shapes) +
scale_x_discrete(limits = axis_grp_order) +
scale_y_log10(breaks = c(100, 1000, 2000, 3000, 5000, 10000),
label = c("< 100", 1000, 2000, 3000, 5000, 10000)) +
facet_wrap(~visitno) +
labs(y = "IgG binding antibody units (MFI - blank)", x = "") +
theme_classic() +
theme(panel.background = element_rect(color="black"), legend.position = "bottom")Response magnitude box plots - version 2
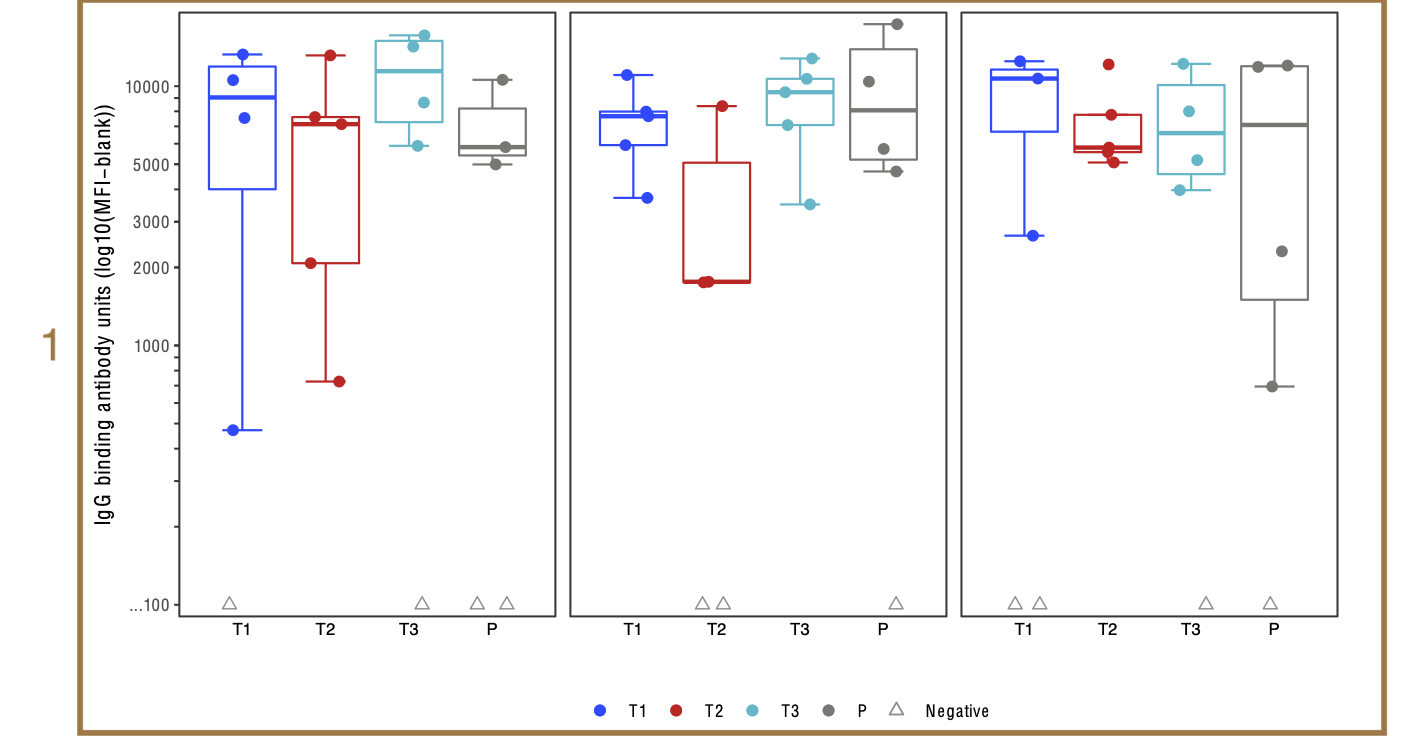
- To do before final figure: tinkering with
theme().- Remove panel (strip) labels for stacking.
strip.text = element_blank() - Wrap individual plots with boxes.
panel.background = element_rect(color="black").
- Remove panel (strip) labels for stacking.
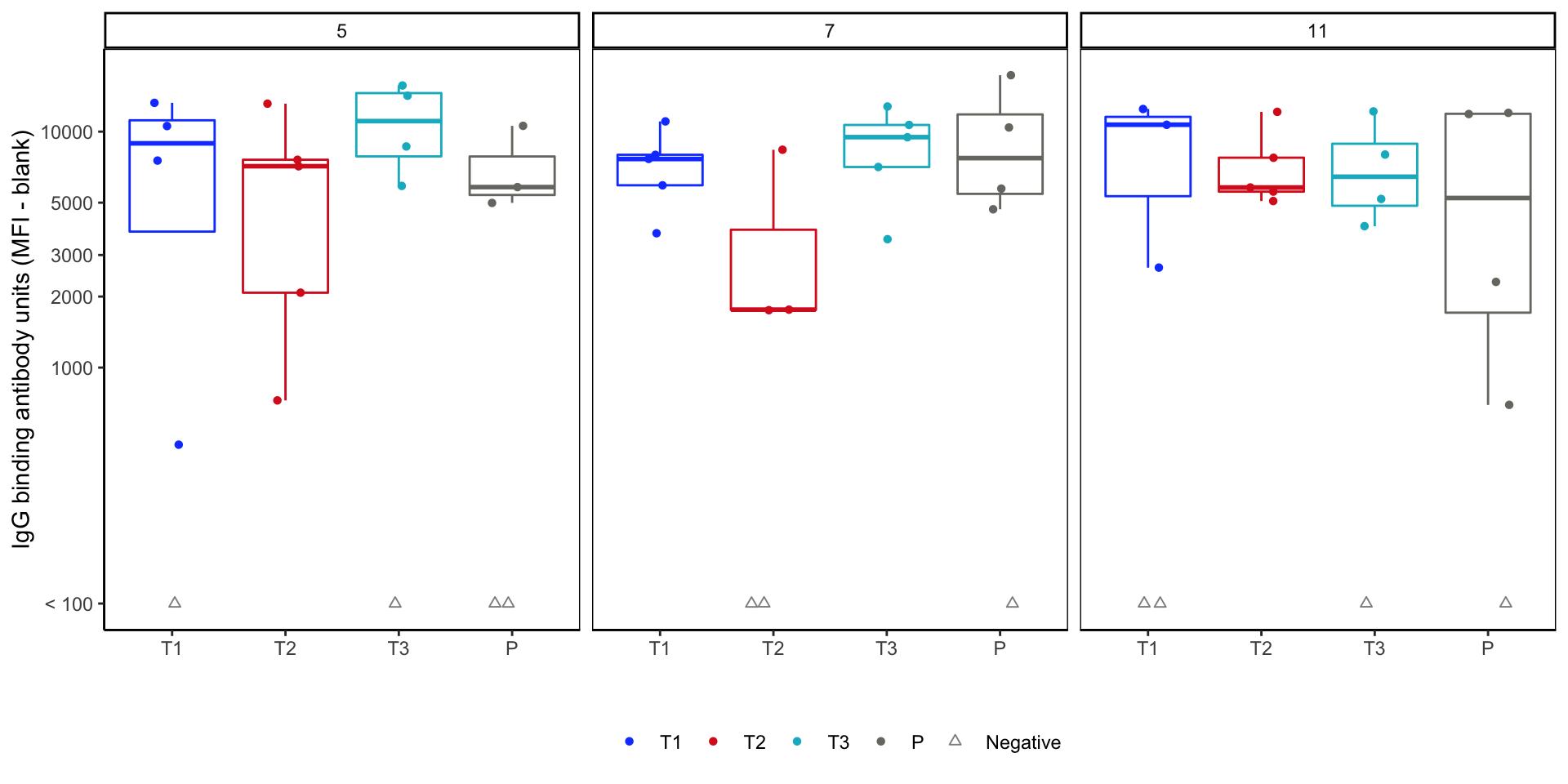
Finalized magnitude figure
Finishing HVTN Example
Walkthrough parts 2 and 3. Code is in the worksheet.

Responder total tally - step 1
Approach: Summarize the data and get the text aligned on a plot.
response_rate_summary = mock_bama_example %>%
group_by(rx_code, antigen, visitno) %>%
summarize(
Total = n(),
Responders = sum(response),
rr = mean(response),
.groups = "drop"
)
rr_txt_pl_skeleton = ggplot(response_rate_summary,
aes(x = rx_code, y = 1,
label = paste(Responders, Total, sep = "\n"))) +
geom_text(size = 3) +
scale_x_discrete(limits = axis_grp_order) +
scale_y_continuous(breaks = 1, labels = "Responders\nTotal", expand = c(0, 0)) +
facet_wrap(~visitno) +
labs(y = "")
rr_txt_pl_skeletonResponder total tally - step 1
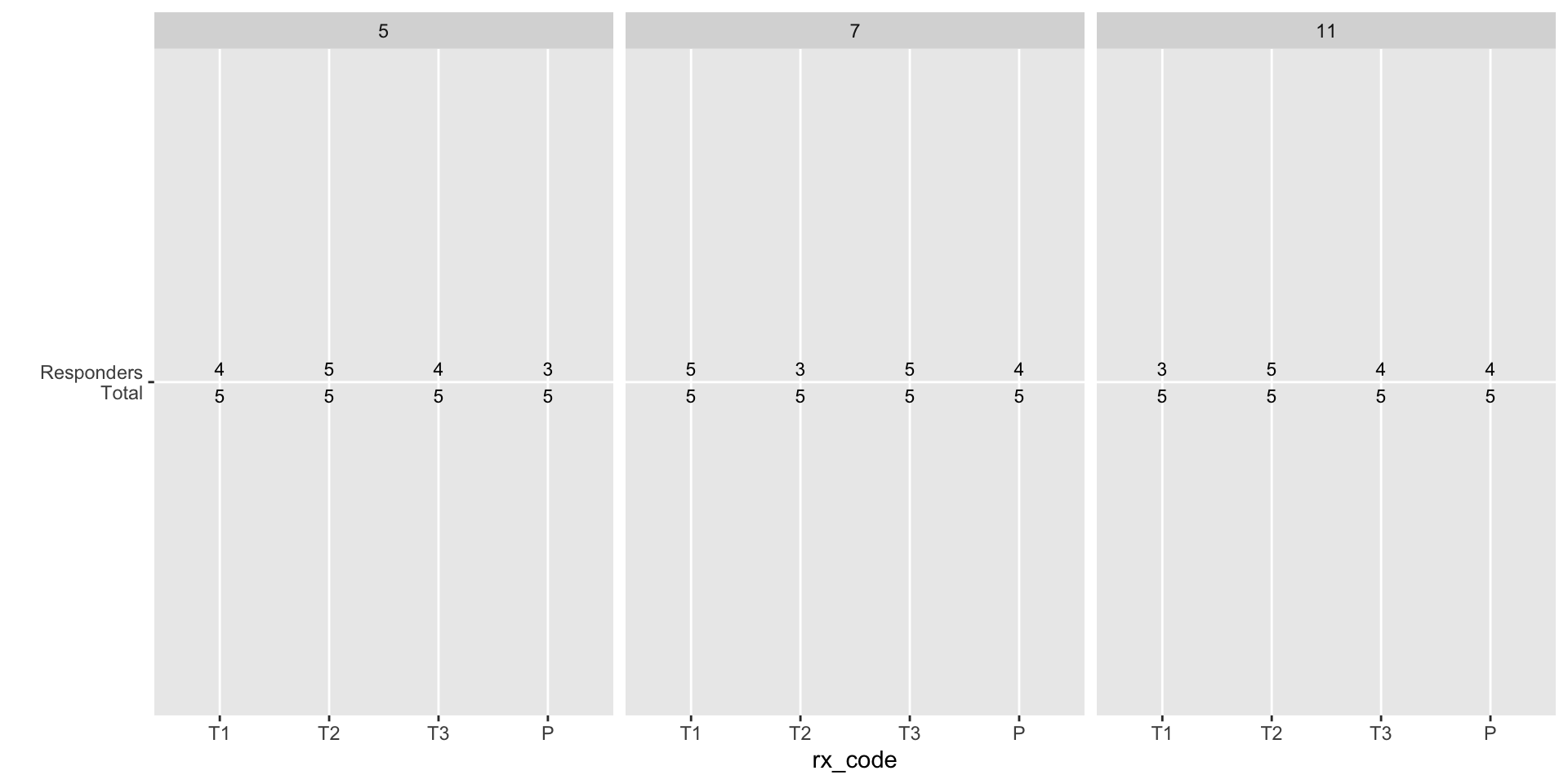
Responder total tally - finalize
Use theme() to remove most plot elements. The axes coordinates are invisible but remain aligned with group.
rr_txt_pl = rr_txt_pl_skeleton +
theme(panel.background = element_blank(), axis.text.x = element_blank(),
axis.title.x = element_blank(), axis.ticks = element_blank(),
strip.background = element_blank(), strip.text = element_blank(),
axis.text.y = element_text(size = 10), plot.margin = margin())
rr_txt_plResponder total tally - finalize
Response rate barplot
- Two potential approaches (both in worksheet):
- Use
stat_summary_binapplied to the full dataset. - Use summarized data from tallies and
geom_bar(stat = 'identity')(see worksheet).
- Use
resp_rate1 = ggplot(mock_bama_example, aes(x = rx_code, y = 100*response, fill = rx_code)) +
stat_summary_bin(fun = "mean", geom = "bar") +
scale_fill_manual(values = hvtn_pl_colors, guide = "none") +
scale_x_discrete(limits = axis_grp_order) +
scale_y_continuous(expand = c(0, 0), limits = c(0, 102)) +
labs(y = "Response rate (%)", x = "") +
facet_wrap(~visitno, labeller =
as_labeller(c(`5` = "Visit 5",
`7` = "Visit 7",
`11` = "Visit 11"))) +
theme_classic() +
theme(strip.background = element_blank(), panel.background = element_rect(color="black"),
plot.title = element_text(size = 12)) +
ggtitle("HVTN Trial IgG Antibody Responses to ag_b")
resp_rate1 Response rate barplot
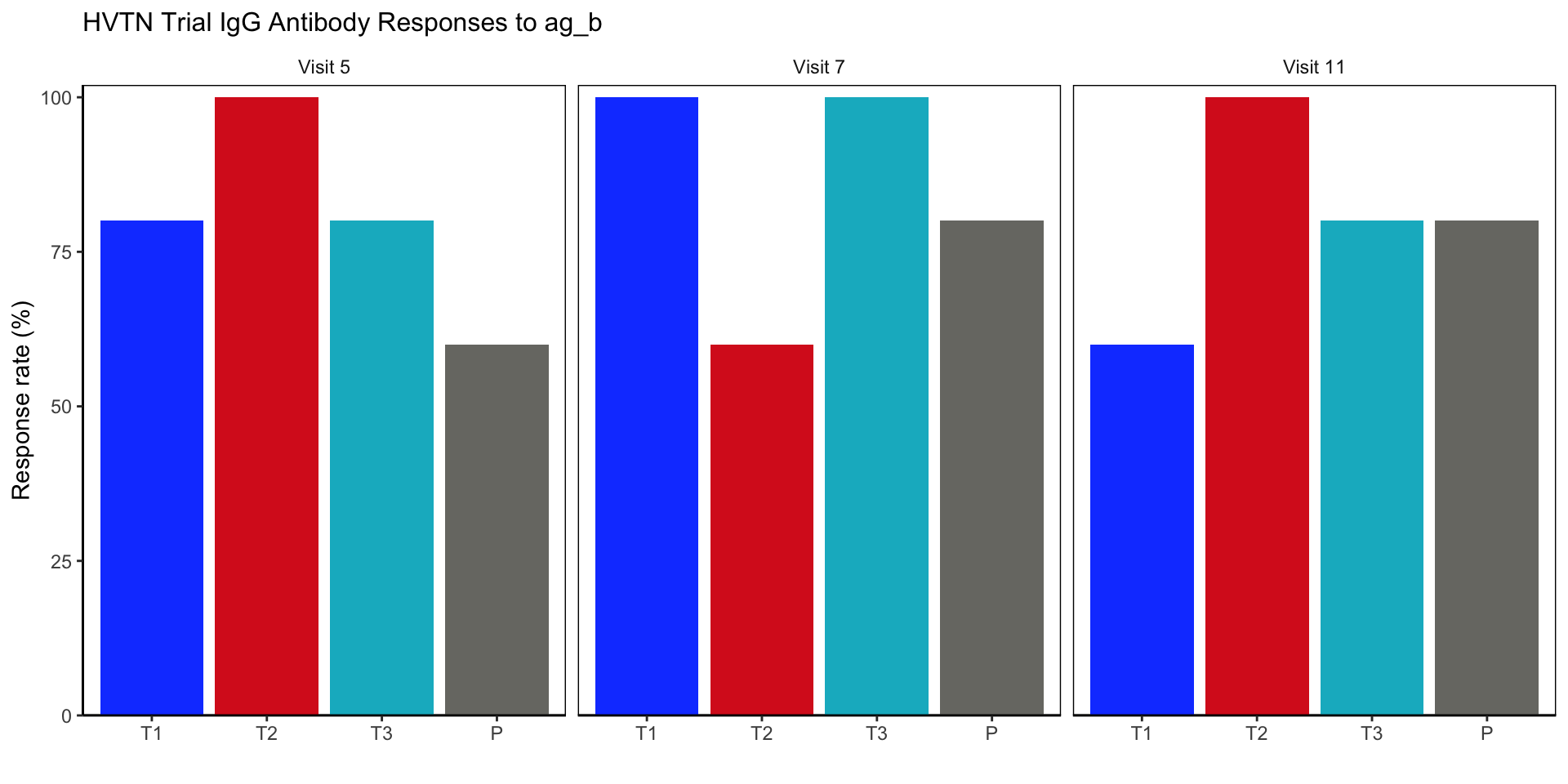
Final Figure
- Use
cowplot::plot_grid
Final Figure

Extra slides
V1 Plot Answer
ggplot(data = mock_bama_example, aes(x = rx_code, y = pmax(100, delta),
color = rx_code)) +
geom_boxplot(data = subset(mock_bama_example, response == 1),
show.legend = F,
outlier.colour = NA) +
geom_point(aes(shape = factor(response), group = rx_code),
position = position_jitter(width = 0.2)) +
scale_colour_manual("", values = hvtn_pl_colors) +
scale_shape_manual("", values = c(2, 16)) +
scale_x_discrete(limits = axis_grp_order) +
scale_y_log10(breaks = c(100, 1000, 2000, 3000, 5000, 10000)) +
facet_wrap(~ visitno) +
labs(y = "IgG binding antibody units (MFI - blank)", x = "") +
theme_classic()